In this blog, we take a closer look at advancements in omics, modernization and large-scale cloud migration of omics ecosystem using AWS Services, and getting started with SourceFuse, an AWS Healthcare Competency Partner.
Omics is an informal term to describe various branches of biological science in healthcare and life sciences (HLS) that end in ‘-omics’, such as genomics, proteomics, metabolomics, metagenomics, phenomics, and transcriptomics. As HLS organizations continue to improve their understanding of omics, and applications in ‘precision medicine’ to enhance personalized care, the global adoption of next-generation sequencing (NGS) techniques by research organizations is rapidly accelerating.
In the era of modern medicine with the advancement of omics findings, the one-size-fits-all therapeutic model is no longer relevant in clinical practice. By leveraging molecular techniques such as NGS, our ultimate goal should be to improve individualized patient care. While sequencing costs continue to decrease, resource and cost constraints associated with compute and storage have grown. In addition, data throughput and more data-heavy applications, such as precision oncology and immuno-therapeutics, have increased.
HLS organizations generate large volumes of clinical data from high throughput genomics sequencers and other clinical devices. These organizations need cost-effective, centrally stored data for applications in distributed locations, to access data for content management, analytics, or processing. The Global Alliance for Genomics and Health (GA4GH) goes further, stating its main objective as: “The responsible and secure sharing of genomic and health data is key to accelerating research and improving human health”.
Read our recent blog: A Data-Driven Future: What’s coming for us?
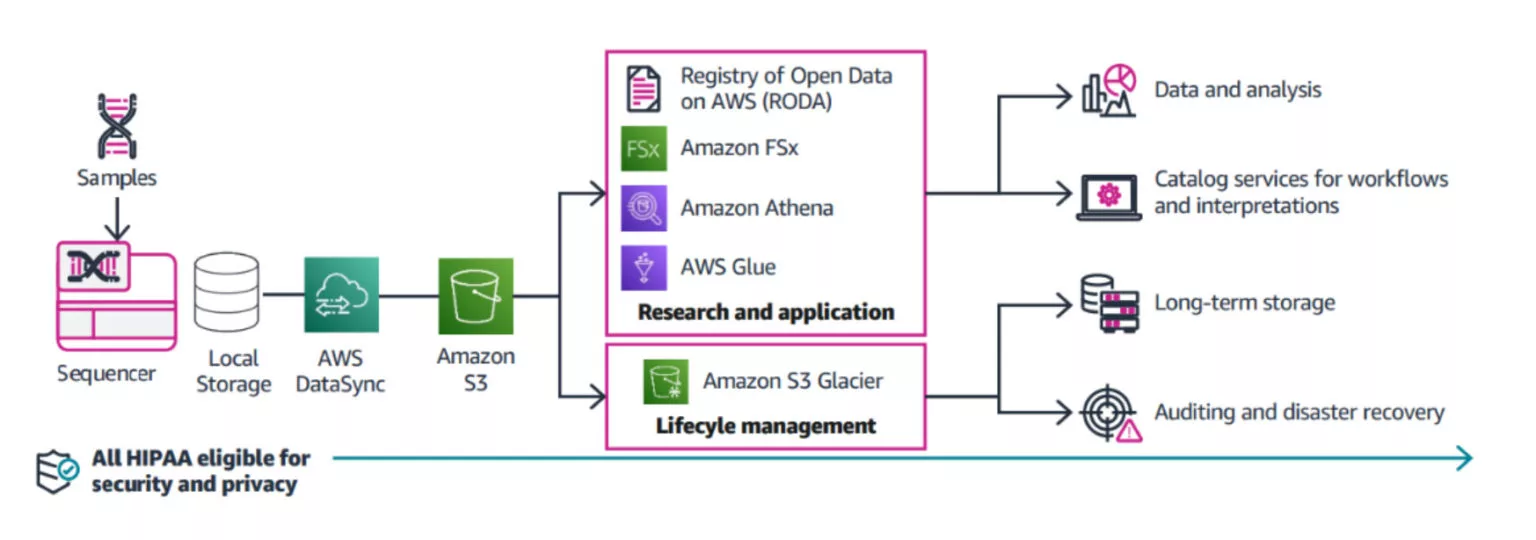
Processing Omics Data
Raw omics data is typically processed through a series of steps, converting it to a format ready for analysis. Each step could have different compute and memory requirements; some steps could be as simple as adding a set of annotations, or as computationally intensive as aligning raw reads to a reference genome. The goal at this stage is to process the data in a cost-effective, scalable, efficient, consistent, and reproducible manner across large datasets.
Once the data is processed, the next step is to query and mine omics data to reveal actionable insights such as discovering new biomarkers or drug targets. At this stage, the goal is to prepare these large datasets so they can be easily queried and, in an interactive manner, answer relevant scientific questions. These datasets could also be used to build complex machine learning (ML) models to analyze population or disease-specific datasets.
Cloud Modernization and Migration Strategy
While there are several migration strategies, the two most common migration methods amongst omics organizations are ‘lift-and-shift’ and ‘modernization-first’. Often, we see life sciences companies select different large-scale cloud migration strategies across different areas of their value chain, depending on their business goals and needs.
Amazon Web Services (AWS) offer hybrid cloud solutions to connect on-premises applications and systems to cloud storage to help reduce costs, minimize management burden, and innovate with your data. For example:
- AWS Outposts Family that enables life sciences organizations to run AWS infrastructure and services on-premises to create a consistent hybrid experience.
- AWS Storage Gateway to seamlessly connect and extend their on-premises application to AWS Storage. This is a prime method for replacing tape libraries with cloud storage, providing cloud storage-backed file shares, or creating a low-latency cache to access data in AWS for on-premises applications.
- AWS Direct Connect that provides a dedicated physical connection, to further accelerate network transfers between on-premises data centers and AWS data centers.
Transfer and Storage of Data from Sequencer to Cloud, Secondary Analysis, Tertiary Analysis and ML
AWS offers options for transferring sequencing data from on-premises storage into an Amazon S3 bucket for storage and downstream analysis. Once a sequencing run has concluded, data can be written to a local storage system folder for transfer to an Amazon S3 bucket using AWS DataSync. For annotation and clinical data files, research scientists and clinical researchers can transfer data to the same S3 bucket using AWS Transfer Family by FTP, SFTP, or FTPS.
The SourceFuse Focus on Healthcare
Data is the most valuable asset of any organization. And with healthcare data subject to the most stringent privacy and protection regulations, your data needs to be handled with extreme care. This is the reason why SourceFuse is focussed on ensuring data security and adherence to regulatory compliance across the healthcare ecosystem.
Having over 16 years of experience in the HLS industry and deep technical expertise, SourceFuse understands the demands and needs of HLS business leaders. Being the first AWS Advanced Consulting Partner in the APAC region and the 15th worldwide to achieve the AWS Healthcare Competency, SourceFuse incorporates advanced AWS tools and services to deliver HIPAA and GDPR compliant solutions.
Being an Amazon HealthLake Partner, SourceFuse supports HLS organizations to achieve the end-goal of healthcare data analytics unlocking powerful insights from omics data and observing health population trends.